Fragment screening is a powerful approach to identifying ligands, and can identify tractable starting points for a wide range of targets. However, fragment screening by conventional biophysical methods typically requires a large amount of protein, takes significant time to perform, and the solubility requirements can restrict the chemical space coverage of the fragment library.
In collaboration with HitGen, we have developed a new approach to fragment screening which exploits DNA encoded methods in order to overcome some of these limitations. Each library member has an individual DNA tag and consists of a fragment linked to a photoactivatable group (usually diazirine). In the screen, photoactivation covalently attaches any binding fragments to the protein; after extensive washing, hits are then identified by PCR and Next Generation Sequencing (NGS).
The sensitivity of DNA-encoding means that only small amounts of protein are required, the screen is rapid and is run at low concentrations. Additionally, the fragments are solubilised by the DNA tag, overcoming the usual requirement for highly soluble compounds. The approach is described in a recent paper, Ma, H. et al., RSC Med. Chem., 2022, 13, 1341 , demonstrating successful identification of fragment hits for a conventional enzyme target, PAK4 kinase and a more challenging and as yet undrugged target, 2-epimerase.
The target protein with an appropriate tag is incubated with the PAC-FragmentDEL library, followed by photoactivation. The protein is then immobilised on a bead and successive wash cycles remove any unbound fragments to leave only covalently bound fragment hits. These are subsequently identified through PCR and sequencing. Control samples are also prepared which are not exposed to UV and those which do not contain target protein.
As with conventional DEL, some form of competitive or comparative screen is typically required. The experiments described in this paper used a competitor compound to identify fragments that bound to a particular site. Alternate strategies include using a peptide, partner protein or mutations to block the site.
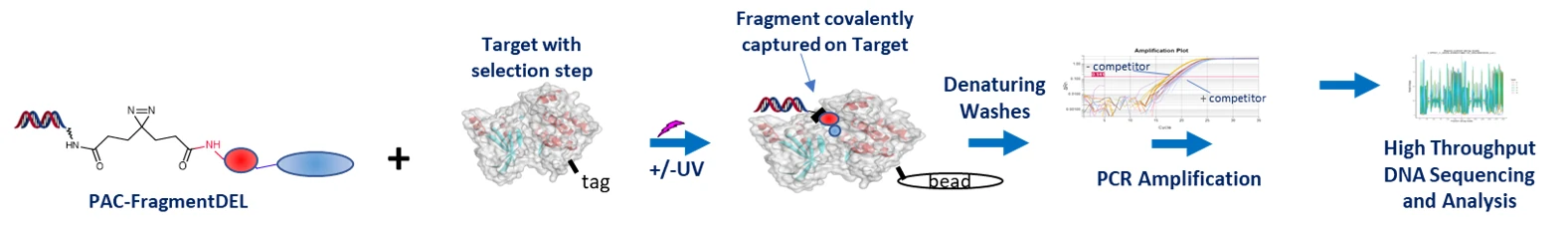
The sequencing output is analysed to identify putative hits- these are prepared without the DNA tag (“off-DNA”) and are subsequently validated by orthogonal biophysical techniques such as surface plasmon resonance (SPR), ligand observed NMR (LO-NMR) and crystallography.
Additionally, the linkage to the DNA provides information about the orientation of the fragment in the binding site, and the validated fragments can be further elaborated through the linker vector to allow rapid exploration of chemical space.
Image: Crystal structure of fragment hit in the ATP-binding site of PAK4 (SPR KD 15μM)
Library members are generated using “split and pool” combinatorial synthesis, with the same fragment headgroups being presented on a variety of linker lengths. This allows the exploration of the depth of the binding site and access to sites which might be sterically hindered. Each individual library member is tagged with a unique DNA code for identification.
In an example library of 70K compounds, 2 library types were generated. The Type 1 library incorporates an extension linker; the Type 2 library is created by merging two smaller fragments in the absence of an extension linker, resulting in larger compounds, which may be advantageous in increasing the scope for protein targets which bind with lower ligand efficiency.
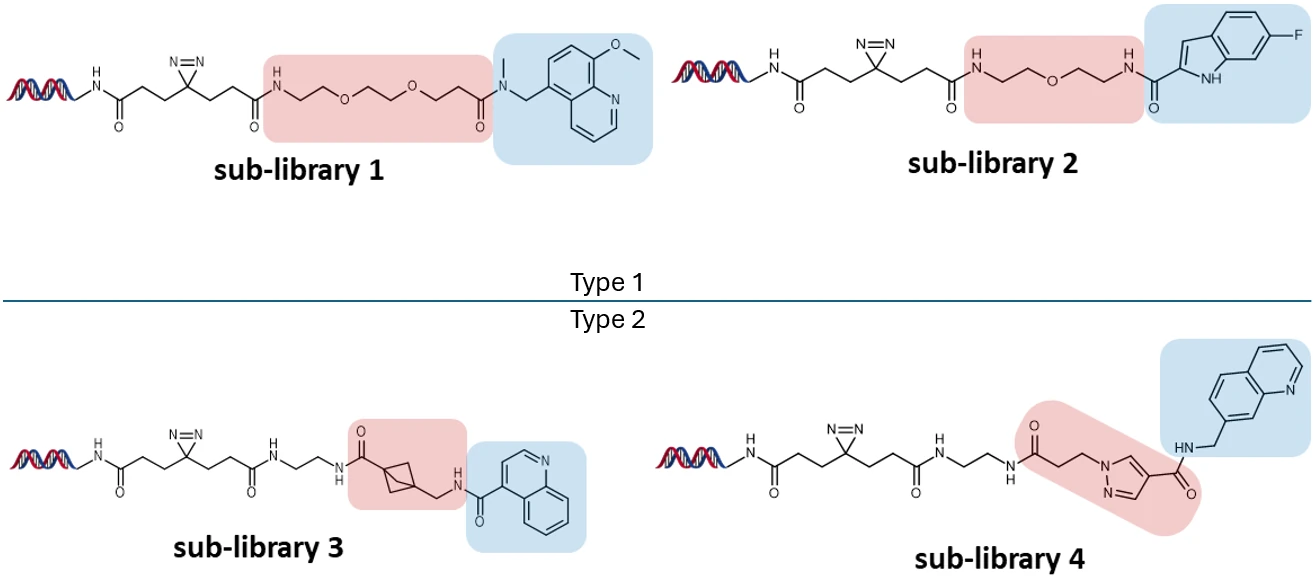
Type 1 and 2 PAC-FragmentDEL Libraries